 Remoscope: a label-free imaging cytometer for malaria diagnostics Trans R Soc Trop Med Hyg 2025
|
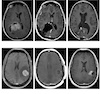 IL-6 underlies microenvironment immunosuppression and resistance to therapy in glioblastoma bioRxiv, 2025
|
 A Trypanosoma cruzi Trans-Sialidase Peptide Demonstrates High Serological Prevalence Among Infected Populations Across Endemic Regions of Latin America medRxiv, 2025
|
 Proteomic profiling of the local and systemic immune response to pediatric respiratory viral infections American Society for Microbiology, 2025
|
 Endogenous antigens shape the transcriptome and TCR repertoire in an autoimmune arthritis model The Journal of Clinical Investigation, 2024
|
 Unveiling the proteome-wide autoreactome enables enhanced evaluation of emerging CAR T cell therapies in autoimmunity J Clin Invest 2024
|
 Total Syntheses of Cyclomarin and Metamarin Natural Products Organic Letters, 2024
|
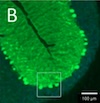 Anti-RGS8 paraneoplastic cerebellar ataxia is preferentially associated with a particular subtype of Hodgkin's lymphoma Springer Nature, 2024
|
 Microbial dynamics and pulmonary immune responses in COVID-19 secondary bacterial pneumonia Nature Portfolio, 2024
|
 Climate, demography, immunology, and virology combine to drive two decades of dengue virus dynamics in Cambodia PNAS, 2024
|
 Molecular mimicry in multisystem inflammatory syndrome in children Nature Portfolio, 2024
|
 Phage Immunoprecipitation-Sequencing Reveals CDHR5 Autoantibodies in Select Patients With Interstitial Lung Disease American College of Rheumatology, 2024
|
|  Please use this chart to choose the ideal size cooler for your shipment, based on number of days in transit. Note that larger samples require larger shipping coolers, because the amount of dry ice will be reduced. |
 This is the protocol that accompanies our 2011 article: Plate-based transfection and culturing technique for genetic manipulation of Plasmodium falciparum, by Florencia Caro and Matt Miller, published in Malaria Journal. Plasmodium plate based transfection protocol |
 This is the raw sequence data that accompanies our 2011 paper Temporal Analysis of the Honey Bee Microbiome Reveals Four Novel Viruses and Seasonal Prevalence of Known Viruses, Nosema, and Crithidia This data was generated on the Illumina platform and consists of a lane of paired end 65nt read files. Both Lake Sinai virus 1 and Lake Sinai virus 2 may be assembled from these reads. The file is large, consisting of a 4Gb zip compressed file. Download raw sequence read archive (Warning: 4Gb zip file) |
 We are pleased to introduce MITOMI 2.0, a microfluidic platform and analysis pipeline for high-throughput measurement of transcription factor DNA sequence preferences and interaction affinities. Using a panel of 28 S. cerevisiae transcription factors, including 2 that were previously uncharacterized, we demonstrated the ability to comprehensively identify both high- and low-affinity target sequences and directly measure relative binding affinities. We hope that both the extensive data set presented here (including affinity information for each transcription factor binding to 1457 oligonucleotide sequences) and future use of this technique help elucidate the fundamental mechanisms by which transcription factors regulate gene expression. Click here to download the raw Mitomi v2 data from the paper. (18Mb zip archive) |
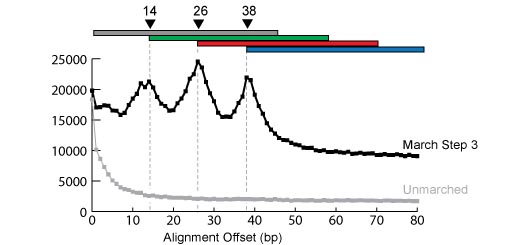 The Long March is a simple technique for extending the amount of coverage with short read sequencers, such as Solexa (Illumina). We have demonstrated the utility of the technique using the malaria transcriptome and serum from a hepatitis B positive patient as an example. Long March Supplemental Data |
 The RhinoBase is a companion website to our publication on selective pressure operating on the human rhinovirus genome. This site contains easy downloads of all the new complete rhinovirus genomes we sequenced, data mapped onto crystal structures, movies, media, and other helpful resources. Visit the RhinoBase |
 The database that accompanies our 2003 PLoS Biology paper features a query interface for viewing the raw data from this 48 hour long experiment. From a synchronized culture of Plasmodium falciparum, samples were withdrawn on the hour, every hour, for the entire intraerythrocytic lifecycle. DeRisi Lab Malaria Transcriptome Database |
 This is the web supplement to our 2002 PNAS manuscript introducing the ViroChip, a microarray platform for the detection and discovery of viral pathogens. This web supplement provides access to primary datasets, raw images, and analysis from the paper. ViroChip Article Web Supplement |
 In eukaryotic cells, cohesin holds sister chromatids together until they separate into daughter cells during mitosis.We have used chromatin immunoprecipitation coupled with microarray analysis (ChIP chip) to produce a genome-wide description of cohesin binding to meiotic and mitotic chromosomes of S. cerevisiae.The genome-wide view of mitotic and meiotic cohesin binding provides an important framework for the exploration of cohesins and cohesion in other genomes. Cohesin Mapping Website (at the Stowers Institute) |
 This is the original website for downloading data, figures, and information associated with mapping of meiotic recombination hotspots. This work was published in PNAS, in 2000. Hotspot Web Supplement |
|
