 Remoscope: a label-free imaging cytometer for malaria diagnostics Trans R Soc Trop Med Hyg 2025
|
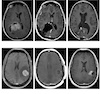 IL-6 underlies microenvironment immunosuppression and resistance to therapy in glioblastoma bioRxiv, 2025
|
 A Trypanosoma cruzi Trans-Sialidase Peptide Demonstrates High Serological Prevalence Among Infected Populations Across Endemic Regions of Latin America medRxiv, 2025
|
 Proteomic profiling of the local and systemic immune response to pediatric respiratory viral infections American Society for Microbiology, 2025
|
 Endogenous antigens shape the transcriptome and TCR repertoire in an autoimmune arthritis model The Journal of Clinical Investigation, 2024
|
 Unveiling the proteome-wide autoreactome enables enhanced evaluation of emerging CAR T cell therapies in autoimmunity J Clin Invest 2024
|
 Total Syntheses of Cyclomarin and Metamarin Natural Products Organic Letters, 2024
|
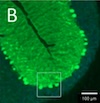 Anti-RGS8 paraneoplastic cerebellar ataxia is preferentially associated with a particular subtype of Hodgkin's lymphoma Springer Nature, 2024
|
 Microbial dynamics and pulmonary immune responses in COVID-19 secondary bacterial pneumonia Nature Portfolio, 2024
|
 Climate, demography, immunology, and virology combine to drive two decades of dengue virus dynamics in Cambodia PNAS, 2024
|
 Molecular mimicry in multisystem inflammatory syndrome in children Nature Portfolio, 2024
|
 Phage Immunoprecipitation-Sequencing Reveals CDHR5 Autoantibodies in Select Patients With Interstitial Lung Disease American College of Rheumatology, 2024
|
|  The DeRisi Lab focuses on genomic approaches to the study of infectious disease. Specifically, we are studying Plasmodium falciparum, the causative agent of the most deadly form of human malaria. We are also involved in a major effort for the discovery of new viral pathogens associated with diseases of unknown etiology. The Research boxes below are taken from past postings on the front page of this website. |
 Despite recent impressive gains over the last decade in the fight against malaria, there exists an urgent need for new, fast acting medications to treat Plasmodium falciparum, the causative agent of the most deadly form of the disease. The need is especially acute given the emergence of artemesinin resistance in Asia. Here, we are please to announce a new candidate for a rapid-acting blood stage anti-malarial compound, SJ733. This compound has won Pre-Clinical Approval with the Malaria Medicines Venture. Its target, PfATP4, a sodium extrusion pump, is shown in a model above, with the compound docked in the presumptive sodium channel, where our data suggests that it impairs the function of the protein. The above image was rendered with UCSF Chimera by J.DeRisi, using a homology model and docked compound generated by J.Horst. Read the Paper Here! |
 Ball Python Nidovirus: a Candidate Etiologic Agent for Severe
Respiratory Disease in Python regius
Ball pythons are popular pets because of their diverse coloration, generally nonaggressive behavior, and relatively
small size. Since the 1990s, veterinarians have been aware of an infectious respiratory disease of unknown cause in ball pythons
that can be fatal. We used unbiased shotgun sequencing to discover a novel virus in the order Nidovirales that was present in
cases but not controls. While nidoviruses are known to infect a variety of animals, this is the first report of a nidovirus recovered
from any reptile. This report will enable diagnostics that will assist in determining the role of this virus in the causation of disease,
which would allow control of the disease in zoos and private collections. Given its evolutionary divergence from known
nidoviruses and its unique host, the study of reptile nidoviruses may further our understanding of related diseases and the viruses
that cause them in humans and other animals.
Photo credit: Christina Wozniak.
[ Read the full open-access article at the mBio website ] |
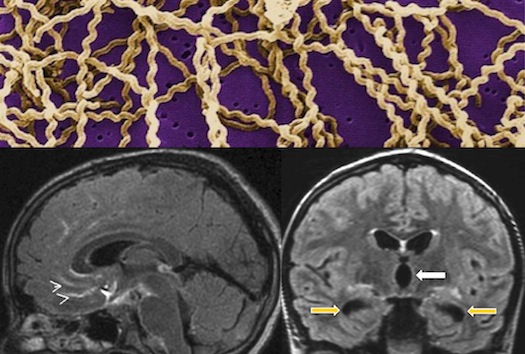 Actionable Diagnosis of Neuroleptospirosis by Next-Generation Sequencing A 14-year-old boy with severe combined immunodeficiency presented 3 times to a medical facility over 4 months with fever and headache that progressed to hydrocephalus and status epilepticus necessitating a medically-induced coma after starting empiric steroids for a suspected autoimmune condition. An exhaustive diagnostic workup including brain biopsy was unrevealing. In collaboration with Dr. Charles Chiu’s lab (UCSF Department of Laboratory Medicine), unbiased next-generation sequencing of the patient’s cerebrospinal fluid and computational analysis of the results using an ultra-rapid bioinformatics pipeline for comprehensive pathogen detection identified 475 sequence reads out of 3,063,784 (0.016%) corresponding to Leptospira spp. This result was obtained within 48 hours of sample receipt. The patient responded dramatically when treated with appropriate antimicrobials and discontinuation of steroids, and was discharged home 24 days later near his premorbid baseline.
[ Read the Article in the New York Times ] •
[ Press Release ]
|
 Malaria caused by Plasmodium spp parasites is a profound human health problem that has shaped our evolutionary past and continues to influence modern day with a disease burden that disproportionately affects the world's poorest and youngest. A plastid organelle, the apicoplast, has been hailed as Plasmodium's Achilles' heel? because it contains bacteria-derived pathways that have no counterpart in the human host and therefore may be ideal drug targets. In this study, we use a simple chemical method to generate parasites that have lost their apicoplast, normally a deadly event, but which survive rescued by the addition of an essential metabolite to the culture. This chemical rescue demonstrates that the apicoplast serves only a single essential function, namely isoprenoid precursor biosynthesis during blood-stage growth, validating this metabolic function as a viable drug target. Moreover, the apicoplast-minus Plasmodium strains generated in this study will be a powerful tool for identifying apicoplast-targeted drugs and as a potential vaccine strain with significant advantages over current vaccine technologies. Read the Full Press Release Here |
 For many years, a mysterious disease in boa constrictors and pythons called Inclusion Body Disease (IBD) has been the scourge of aquariums, breeders, and pet snake owners. The disease is ultimately fatal, and until now, its cause was unknown. Using the latest methods in ultra deep sequencing and de novo sequence assembly, the DeRisi lab, in collaboration with the California Academy of Science and local veterinarians, has identified a novel snake arenavirus that is closely linked to the disease. The paper describing this discovery is published in ASM's new open-access journal mBio, to be released August 14th.
[ The Paper at mBio ] • [ Press Release ] • [ Press Release Video ] • [ FAQ ] |
 A 10-month study of healthy honey bees by University of California, San Francisco scientists has identified four new viruses that infect bees, while revealing that each of the viruses or bacteria previously linked to colony collapse is present in healthy hives as well.
The study followed 20 colonies in a commercial bee-keeping operation of more than 70,000 hives as they traversed the country pollinating crops, to answer one basic question: what does a normal hive look like in terms of viruses and bacteria through the seasons?
View Press Release |
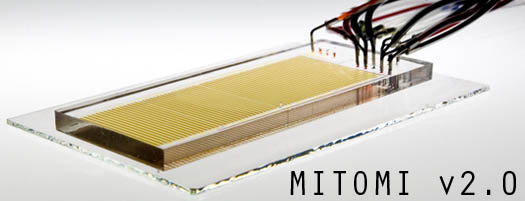 We are pleased to introduce MITOMI v2.0, a microfluidic platform, developed in collaboration with the Quake lab at Stanford, and analysis pipeline for high-throughput measurement of transcription factor DNA sequence preferences and interaction affinities. Using a panel of 28 S. cerevisiae transcription factors, including 2 that were previously uncharacterized, we demonstrated the ability to comprehensively identify both high- and low-affinity target sequences and directly measure relative binding affinities. For more information, click the "More" link. Click here to go to the raw data download. |
 We are pleased to release PRICE (Paired-Read Iterative Contig Extension), a de novo genome assembler implemented in C++. Its name describes the strategy that it implements for genome assembly: PRICE uses paired-read information to iteratively increase the size of pre-assembled contigs. It was designed to address the challenge of assembling viral genomes that constituted a small minority of the reads within ultra-deep, short-read, metagenomic, shotgun datasets. PRICE has already enabled the discovery of several novel virus genomes from such complex datasets, and it is also being applied to the de novo assembly of large individual genomes. It is provided here as a beta release, pre-publication. Download PRICE (beta) |
 We are pleased to release HMMSplicer to the community. This open-source software package discovers splice sites in high throughput sequencing datasets without using gene models. HMMSplicer can also be used to find non-canonical junctions as well. HMMSplicer was benchmarked on publicly available A. thaliana, H. sapiens, and P. falciparum datasets and performs well on all genomes. Information about the datasets tested, including the exact command parameters and the final results, is provided. HMMSplicer is implemented in Python and is freely available for all. Manuscript is in press at PLoS ONE. Learn more and download HMMSplicer here! |
|
