 Remoscope: a label-free imaging cytometer for malaria diagnostics Trans R Soc Trop Med Hyg 2025
|
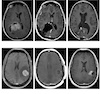 IL-6 underlies microenvironment immunosuppression and resistance to therapy in glioblastoma bioRxiv, 2025
|
 A Trypanosoma cruzi Trans-Sialidase Peptide Demonstrates High Serological Prevalence Among Infected Populations Across Endemic Regions of Latin America medRxiv, 2025
|
 Proteomic profiling of the local and systemic immune response to pediatric respiratory viral infections American Society for Microbiology, 2025
|
 Endogenous antigens shape the transcriptome and TCR repertoire in an autoimmune arthritis model The Journal of Clinical Investigation, 2024
|
 Unveiling the proteome-wide autoreactome enables enhanced evaluation of emerging CAR T cell therapies in autoimmunity J Clin Invest 2024
|
 Total Syntheses of Cyclomarin and Metamarin Natural Products Organic Letters, 2024
|
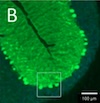 Anti-RGS8 paraneoplastic cerebellar ataxia is preferentially associated with a particular subtype of Hodgkin's lymphoma Springer Nature, 2024
|
 Microbial dynamics and pulmonary immune responses in COVID-19 secondary bacterial pneumonia Nature Portfolio, 2024
|
 Climate, demography, immunology, and virology combine to drive two decades of dengue virus dynamics in Cambodia PNAS, 2024
|
 Molecular mimicry in multisystem inflammatory syndrome in children Nature Portfolio, 2024
|
 Phage Immunoprecipitation-Sequencing Reveals CDHR5 Autoantibodies in Select Patients With Interstitial Lung Disease American College of Rheumatology, 2024
|
|  We are pleased to release HMMSplicer to the community. This open-source software package discovers splice sites in high throughput sequencing datasets without using gene models. HMMSplicer can also be used to find non-canonical junctions as well. HMMSplicer was benchmarked on publicly available A. thaliana, H. sapiens, and P. falciparum datasets and performs well on all genomes. Information about the datasets tested, including the exact command parameters and the final results, is provided. HMMSplicer is implemented in Python and is freely available for all. HMMSplicer was written by Michelle Dimon. Manuscript is in press at PLoS ONE. Learn more and download HMMSplicer here! |
|
